
Bottles by Elevate
Cells make use of tiny machinery known as proteins for different vital processes. They depend on these strings of amino acids for everything from signaling, to regulation, and to catalysis. Since there are countless processes cells are responsible for to keep the organism alive and functional, one would imagine there are an equally immense number of genes present to code these hard-working proteins. What has puzzled scientists for so long however is how unexpectedly short bacterial DNA can be despite this presumption. In addition, typical discovery of new proteins are worked in reverse to identify their respective genes and while many have done so not all proteins have happily given up their genetic codes.
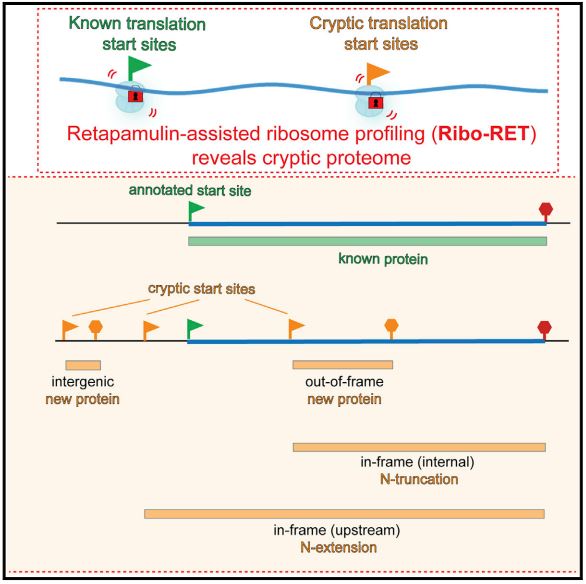
Meydan et. Al. show that a certain antibiotic drug can actually help answer these unsolved mysteries in E. coli. Retapamulin (RET) is an antibiotic that comes from a fungal class of inhibitors which exclusively act on the ribosomes which produce proteins. What makes it so helpful for this line of research is that it selectively inhibits ribosome activity at the start of the protein coding gene, effectively identifying the translation initiation sequence (TIS). Some hypotheses suppose that additional hidden starting sequences known as internal translation initiation sequences (iTIS) may exist. Unlike primary TIS (pTIS) which have been researched extensively and are readily apparent at the start of genes, iTIS are not well understood if at all identified. Proteins made from an iTIS are expected to carry out similar processes as their parent pTIS protein given that they make use of the same code. There are however instances upon which they differ in function when the reading frame shifts and the amino acid codon reading changes.
The group proves that the RET antibiotic stops exclusively at these internal TIS by using toeprinting analysis and ribosome halting data. These techniques made use of ribosome interactions on a piece of mRNA which is meant to be translated into protein. When the ribosome tries to translate the protein, a protein stuck at TISs would prevent further movement and create truncated segments of the proteins. These proteins would be long enough to point back at the TIS which started them. In effect, when RET was given to the E. coli samples the ribosomes were found stuck to specific regions of the mRNA over others. pTIS were used as a control to see if RET did in fact stop ribosomes at the start by comparing the other regions of mRNA where ribosomes were found. The regions where a ribosome was found would essentially be tallied up to create visual bar graph of sorts to see where most were found. If a segment had a lot of ribosomes solely on one part of the mRNA, this was a good suggestion that an iTIS was present. Not only would a TIS be found within a gene and end at the same sequence, but some genes may be found outside the gene, start outside but end with the same sequence, or start and stop within a gene with different starts and stops. This is put simply by the graph presented in their research:
All of this in mind, Meydan and colleagues set out to identify the 239 iTIS in their primary target E. coli BW2543, a particular strand of E. coli. This in and of itself allows for studies to be conducted on the nature of the new alternate proteins hidden within others. They do however also include a comparison with another strain of E. coli BL20 and found 620 iTIS associated with it. Of these, 124 sequences were common to both of the strains suggesting some conservation but also a distinct strain specific need for alternative proteins. They also identify specific genes such as arcB and speA which show ribosomal peaks for iTIS. While arcB may allow for heterodimer formation, speA iTIS forms two distinct isoforms. The isoforms are analyzed and found to be 74 and 70 kDa where the former has a specific secretionary segment at the beginning allowing it to leave the cell’s cytoplasm. This is an example of alternative proteins with similar functions to the primary protein. In this case the speA alternative lacks a 4 kDa secretion sequence at the beginning which limits it to staying in the cell’s cytoplasm only.
Using this newly developed Ribo-RET sequencing technique, it is possible to find new alternative proteins coded in the genome. This may help explain the small sized genomes of bacteria which need to code for multitudes of proteins necessary for their survival. Due to the effectiveness of the Ribo-RET sequencing, previously discovered proteins with no known genes may find their code amongst the newly discovered iTIS. This will prove useful to properly understand their function and use, and allow a new handle for scientists to re-purpose these bio-machines. With this research we can really see how bacterial alternative proteins are just as crafty as alternative splicing in eukaryotic cells.
In Depth
This research may prove to be very impactful not only for future and past research but may also change the way we look at bacterial genomes. A lot of research can follow up on this paper to flush out those details and identify proteins or new gene sequences. The research paper has a lot of great diagrams and figures which can simplify the reading comprehension process. Methods are detailed and can help understand what steps were taken. There were good additional steps made to identify protein gene sequences for known proteins. The comparisons across strains of E. coli is yet another facet of this research which presents interesting data. Evolutionary studies may be able to make use of the fact that some new TIS are conserved across subspecies and perhaps even species. This may help statistical analysis of evolutionary trees as well as understanding the evolution of different genes. Overall this study is well organized and has helpful supporting figures, all of which present a strong step in a new direction.
Meydan, S., Marks, J., Klepacki, D., Sharma, V., Baranov, P. V., Firth, A. E., … & Mankin, A. S. (2019). Retapamulin-assisted ribosome profiling reveals the alternative bacterial proteome. Molecular Cell.
Elevate. https://unsplash.com/@elevatebeer
Unsplash. http://unsplash.com

You must be logged in to post a comment.